Last year the artificial intelligence group DeepMind cracked a mystery that has flummoxed scientists for decades: stripping bare the structure of proteins, the building blocks of life. Now, having amassed a database of nearly all human protein structures, the company is making the resource available online free for researchers to use.
The key to understanding our basic biological machinery is its architecture. The chains of amino acids that comprise proteins twist and turn to make the most confounding of 3D shapes. It is this elaborate form that explains protein function; from enzymes that are crucial to metabolism to antibodies that fight infectious attacks.
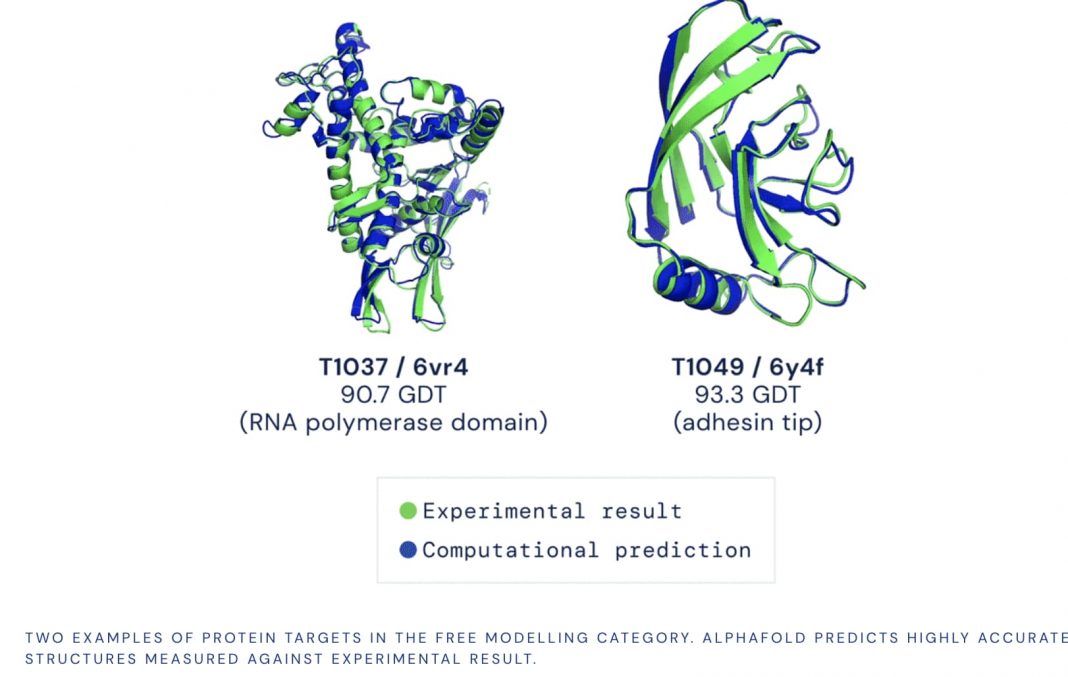
Despite years of onerous and expensive lab work that began in the 1950s, scientists have only decoded the structure of a fraction of human proteins. DeepMind’s AI program, AlphaFold, has predicted the structure of nearly all 20,000 proteins expressed by humans. In an independent benchmark test that compared predictions to known structures, the system was able to predict the shape of a protein to a good standard 95% of time.
DeepMind, which has partnered with the European Molecular Biology Laboratory’s European Bioinformatics Institute (EMBL-EBI), hopes the database will help researchers to analyse how life works at an atomic scale by unpacking the apparatus that drives some diseases, make strides in the field of personalised medicine, create more nutritious crops and develop “green enzymes” that can break down plastic.
Collaboration in recent months with scientists working on a range of projects – from diseases that disproportionately affect poorer parts of the world to studying antibiotic resistance or the biology of the virus that causes Covid – has already begun.
“The applications are actually limited only by our imagination – but at a more fundamental level, the AlphaFold database will increase our understanding of how proteins function, and their role in the fundamental processes of life,” said Prof Edith Heard, the director-general of the EMBL.
“This understanding means we can be better equipped to unravel the molecular mechanisms of life and accelerate our pursuits to protect and treat human health, as well as the health of our planet, and making this tool open access will accelerate the power of research discovery and innovation for scientists around the world.”
AlphaFold’s ability to predict protein structure with dizzying accuracy was unveiled at the biennial “protein olympics” last year. Participants were given the amino acid sequences for about 100 proteins and challenged to work them out. AlphaFold not only eclipsed the performance of other computer programs but achieved accuracy analogous to laborious lab-based methods.
“I almost fell off my chair in just excitement and amazement that this longstanding problem of how proteins fold had been solved,” said Prof Ewan Birney, the director of the EMBL-EBI, after the results were first presented in November.
“This dataset is rather like the human genome … and it’s this dataset where we start some new bits of science that we weren’t able to do beforehand. I’m very excited to start walking down that road.”